Models
Model benchmarks for classifiers are here
Model Interface
- class torchxrayvision.models.Model
The library is composed of core and baseline classifiers. Core classifiers are trained specifically for this library and baseline classifiers come from other papers that have been adapted to provide the same interface and work with the same input pixel scaling as our core models. All models will automatically resize input images (higher or lower using bilinear interpolation) to match the specified size they were trained on. This allows them to be easily swapped out for experiments. Pre-trained models are hosted on GitHub and automatically downloaded to the user’s local ~/.torchxrayvision directory.
Core pre-trained classifiers are provided as PyTorch Modules which are fully differentiable in order to work seamlessly with other PyTorch code.
- forward(x: Tensor) Tensor
The model will output a tensor with the shape [batch, pathologies] which is aligned to the order of the list model.pathologies.
preds = model(img) print(dict(zip(model.targets, preds.tolist()[0]))) # {'Atelectasis': 0.5583771, # 'Consolidation': 0.5279943, # 'Infiltration': 0.60061914, # ...
- targets: List[str]
Each classifier provides a field model.targets which aligns to the list of predictions that the model makes. Depending on the weights loaded this list will change. The predictions can be aligned to pathology names as follows:
- features(x: Tensor) Tensor
The pre-trained models can also be used as features extractors for semi-supervised training or transfer learning tasks. A feature vector can be obtained for each image using the model.features function. The resulting size will vary depending on the architecture and the input image size. For some models there is a model.features2 method that will extract features at a different point of the computation graph.
feats = model.features(img)
XRV Pathology Classifiers
- class torchxrayvision.models.DenseNet(weights=SPECIFY, op_threshs=None, apply_sigmoid=False)
Based on “Densely Connected Convolutional Networks”
Possible weights for this class include:
## 224x224 models model = xrv.models.DenseNet(weights="densenet121-res224-all") model = xrv.models.DenseNet(weights="densenet121-res224-rsna") # RSNA Pneumonia Challenge model = xrv.models.DenseNet(weights="densenet121-res224-nih") # NIH chest X-ray8 model = xrv.models.DenseNet(weights="densenet121-res224-pc") # PadChest (University of Alicante) model = xrv.models.DenseNet(weights="densenet121-res224-chex") # CheXpert (Stanford) model = xrv.models.DenseNet(weights="densenet121-res224-mimic_nb") # MIMIC-CXR (MIT) model = xrv.models.DenseNet(weights="densenet121-res224-mimic_ch") # MIMIC-CXR (MIT)
- Parameters:
weights – Specify a weight name to load pre-trained weights
cache_dir – Override where the weights will be stored (default is ~/.torchxrayvision/)
op_threshs – Specify a weight name to load pre-trained weights
apply_sigmoid – Apply a sigmoid
- targets: List[str] = ['Atelectasis', 'Consolidation', 'Infiltration', 'Pneumothorax', 'Edema', 'Emphysema', 'Fibrosis', 'Effusion', 'Pneumonia', 'Pleural_Thickening', 'Cardiomegaly', 'Nodule', 'Mass', 'Hernia', 'Lung Lesion', 'Fracture', 'Lung Opacity', 'Enlarged Cardiomediastinum']
- class torchxrayvision.models.ResNet(weights=SPECIFY, op_threshs=None, apply_sigmoid=False)
Based on “Deep Residual Learning for Image Recognition”
Possible weights for this class include:
# 512x512 models model = xrv.models.ResNet(weights="resnet50-res512-all")
- Parameters:
weights – Specify a weight name to load pre-trained weights
cache_dir – Override where the weights will be stored (default is ~/.torchxrayvision/)
op_threshs – Specify a weight name to load pre-trained weights
apply_sigmoid – Apply a sigmoid
- targets: List[str] = ['Atelectasis', 'Consolidation', 'Infiltration', 'Pneumothorax', 'Edema', 'Emphysema', 'Fibrosis', 'Effusion', 'Pneumonia', 'Pleural_Thickening', 'Cardiomegaly', 'Nodule', 'Mass', 'Hernia', 'Lung Lesion', 'Fracture', 'Lung Opacity', 'Enlarged Cardiomediastinum']
XRV ResNet Autoencoder
- class torchxrayvision.autoencoders.ResNetAE(weights=SPECIFY)
A ResNet based autoencoder.
Possible weights for this class include:
“101-elastic” trained on PadChest, NIH, CheXpert, and MIMIC. From the paper https://arxiv.org/abs/2102.09475
ae = xrv.autoencoders.ResNetAE(weights="101-elastic") # trained on PadChest, NIH, CheXpert, and MIMIC z = ae.encode(image) image2 = ae.decode(z)
- params:
weights (str): Weights to use. See above for options. cache_dir (str): Override directory used to store cached weights (default: ~/.torchxrayvision/)
CheXpert Pathology Classifier
- class torchxrayvision.baseline_models.chexpert.DenseNet(weights_zip='', num_models=30)
CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. Irvin, J., et al (2019). AAAI Conference on Artificial Intelligence. http://arxiv.org/abs/1901.07031
Setting num_models less than 30 will load a subset of the ensemble.
Modified for TorchXRayVision to maintain the pytorch gradient tape and also to provide the features() argument.
Weights can be found: https://academictorrents.com/details/5c7ee21e6770308f2d2b4bd829e896dbd9d3ee87 https://archive.org/download/torchxrayvision_chexpert_weights/chexpert_weights.zip
- targets: List[str] = ['Atelectasis', 'Cardiomegaly', 'Consolidation', 'Edema', 'Effusion']
JF Healthcare Pathology Classifier
- class torchxrayvision.baseline_models.jfhealthcare.DenseNet(apply_sigmoid=True)
A model trained on the CheXpert data
https://github.com/jfhealthcare/Chexpert Apache-2.0 License
@misc{ye2020weakly, title={Weakly Supervised Lesion Localization With Probabilistic-CAM Pooling}, author={Wenwu Ye and Jin Yao and Hui Xue and Yi Li}, year={2020}, eprint={2005.14480}, archivePrefix={arXiv}, primaryClass={cs.CV} }
- targets: List[str] = ['Cardiomegaly', 'Edema', 'Consolidation', 'Atelectasis', 'Effusion']
ChestX-Det Segmentation
- class torchxrayvision.baseline_models.chestx_det.PSPNet(cache_dir: str = None)
ChestX-Det Segmentation Model
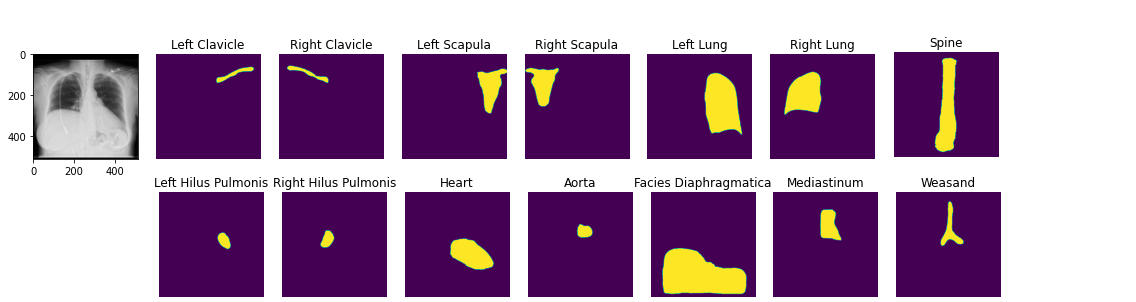
You can load pretrained anatomical segmentation models. Demo Notebook
seg_model = xrv.baseline_models.chestx_det.PSPNet() output = seg_model(image) output.shape # [1, 14, 512, 512] seg_model.targets # ['Left Clavicle', 'Right Clavicle', 'Left Scapula', 'Right Scapula', # 'Left Lung', 'Right Lung', 'Left Hilus Pulmonis', 'Right Hilus Pulmonis', # 'Heart', 'Aorta', 'Facies Diaphragmatica', 'Mediastinum', 'Weasand', 'Spine']
https://github.com/Deepwise-AILab/ChestX-Det-Dataset
@article{Lian2021, title = {{A Structure-Aware Relation Network for Thoracic Diseases Detection and Segmentation}}, author = {Lian, Jie and Liu, Jingyu and Zhang, Shu and Gao, Kai and Liu, Xiaoqing and Zhang, Dingwen and Yu, Yizhou}, doi = {10.48550/arxiv.2104.10326}, journal = {IEEE Transactions on Medical Imaging}, url = {https://arxiv.org/abs/2104.10326}, year = {2021} }
- params:
cache_dir (str): Override directory used to store cached weights (default: ~/.torchxrayvision/)
- targets: List[str] = ['Left Clavicle', 'Right Clavicle', 'Left Scapula', 'Right Scapula', 'Left Lung', 'Right Lung', 'Left Hilus Pulmonis', 'Right Hilus Pulmonis', 'Heart', 'Aorta', 'Facies Diaphragmatica', 'Mediastinum', 'Weasand', 'Spine']
Emory HITI Race
- class torchxrayvision.baseline_models.emory_hiti.RaceModel
This model is from the work below and is trained to predict the patient race from a chest X-ray. Public data from the MIMIC dataset is used to train this model. The native resolution of the model is 320x320. Images are scaled automatically.
model = xrv.baseline_models.emory_hiti.RaceModel() image = xrv.utils.load_image('00027426_000.png') image = torch.from_numpy(image)[None,...] pred = model(image) model.targets[torch.argmax(pred)] # 'White'
@article{Gichoya2022, title = {AI recognition of patient race in medical imaging: a modelling study}, author = {Gichoya, Judy Wawira and Banerjee, Imon and Bhimireddy, Ananth Reddy and Burns, John L and Celi, Leo Anthony and Chen, Li-Ching and Correa, Ramon and Dullerud, Natalie and Ghassemi, Marzyeh and Huang, Shih-Cheng and Kuo, Po-Chih and Lungren, Matthew P and Palmer, Lyle J and Price, Brandon J and Purkayastha, Saptarshi and Pyrros, Ayis T and Oakden-Rayner, Lauren and Okechukwu, Chima and Seyyed-Kalantari, Laleh and Trivedi, Hari and Wang, Ryan and Zaiman, Zachary and Zhang, Haoran}, doi = {10.1016/S2589-7500(22)00063-2}, journal = {The Lancet Digital Health}, pmid = {35568690}, url = {https://www.thelancet.com/journals/landig/article/PIIS2589-7500(22)00063-2/fulltext}, year = {2022} }
- targets: List[str] = ['Asian', 'Black', 'White']
Riken Age Model
- class torchxrayvision.baseline_models.riken.AgeModel
This model predicts age. It is trained on the NIH dataset. The publication reports a mean absolute error (MAE) between the estimated age and chronological age of 3.67 years.
The native resolution of the model is 320x320. Images are scaled automatically.
model = xrv.baseline_models.riken.AgeModel() image = xrv.utils.load_image('00027426_000.png') image = torch.from_numpy(image)[None,...] pred = model(image) # tensor([[50.4033]], grad_fn=<AddmmBackward0>)
Source: https://github.com/pirocv/xray_age
@article{Ieki2022, title = {{Deep learning-based age estimation from chest X-rays indicates cardiovascular prognosis}}, author = {Ieki, Hirotaka et al.}, doi = {10.1038/s43856-022-00220-6}, journal = {Communications Medicine}, publisher = {Nature Publishing Group}, url = {https://www.nature.com/articles/s43856-022-00220-6}, year = {2022} }
- targets: List[str] = ['Age']
Xinario View Model
- class torchxrayvision.baseline_models.xinario.ViewModel
The native resolution of the model is 320x320. Images are scaled automatically.
model = xrv.baseline_models.xinario.ViewModel() image = xrv.utils.load_image('00027426_000.png') image = torch.from_numpy(image)[None,...] pred = model(image) # tensor([[17.3186, 26.7156]]), grad_fn=<AddmmBackward0>) model.targets[pred.argmax()] # Lateral
Source: https://github.com/xinario/chestViewSplit
- targets: List[str] = ['Frontal', 'Lateral']
Mira Sex Model
- class torchxrayvision.baseline_models.mira.SexModel(weights=True)
This model is from the MIRA (Medical Image Representation and Analysis) project and is trained to predict patient sex from a chest X-ray. The model uses a ResNet34 architecture and is trained on CheXpert dataset. The native resolution of the model is 224x224. Images are scaled automatically.
Publication: Algorithmic encoding of protected characteristics in chest X-ray disease detection models B. Glocker, C. Jones, M. Bernhardt, S. Winzeck eBioMedicine. Volume 89, 104467, 2023.
model = xrv.baseline_models.mira.SexModel() image = xrv.utils.load_image('00027426_000.png') image = torch.from_numpy(image)[None,...] pred = model(image) model.targets[torch.argmax(pred)] # 'Male' or 'Female'
@article{MIRA2023, title = {Chexploration: Medical Image Representation and Analysis}, author = {MIRA Team}, journal = {biomedia-mira/chexploration}, url = {https://github.com/biomedia-mira/chexploration}, year = {2023} }
- targets: List[str] = ['Male', 'Female']