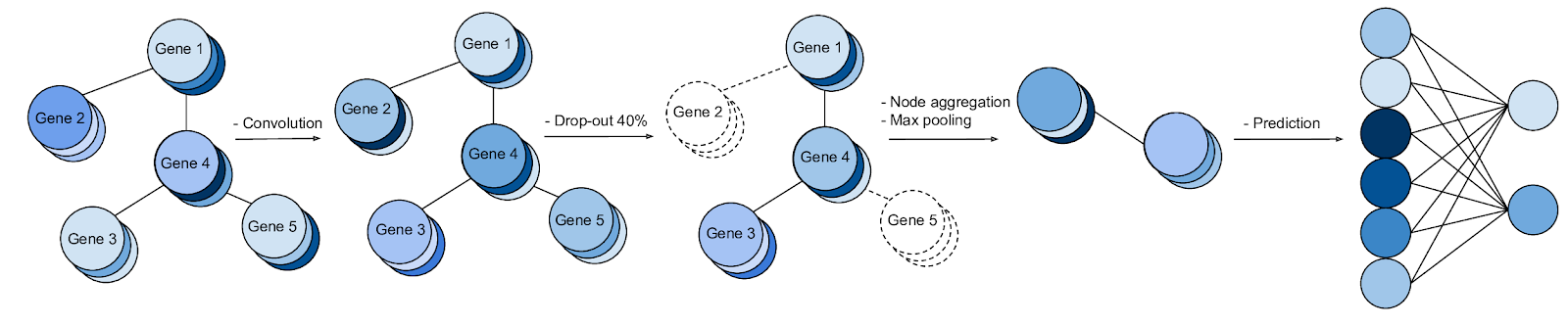
We discuss how gene-gene interaction graphs (same pathway, protein-protein, co-expression, or research paper text association) can be used to impose a bias on a deep neural network model similar to the spatial bias imposed by convolutions on an image. We find this approach provides an advantage for particular tasks in a low data regime but is very dependent on the quality of the graph used.