Motivation: Traditional image attribution methods struggle to satisfactorily explain predictions of neural networks. Prediction explanation is important, especially in medical imaging, for avoiding the unintended consequences of deploying AI systems when false positive predictions can impact patient care. Thus, there is a pressing need to develop improved models for model explainability and introspection.
Specific problem: A new approach is to transform input images to increase or decrease features which cause the prediction. However, current approaches are difficult to implement as they are monolithic or rely on GANs. These hurdles prevent wide adoption.
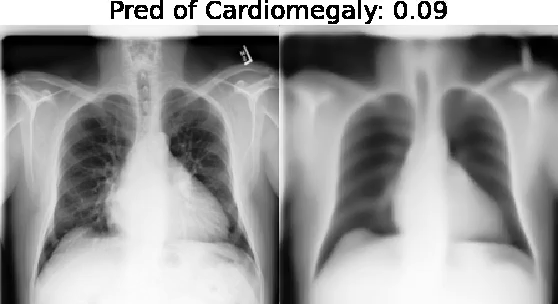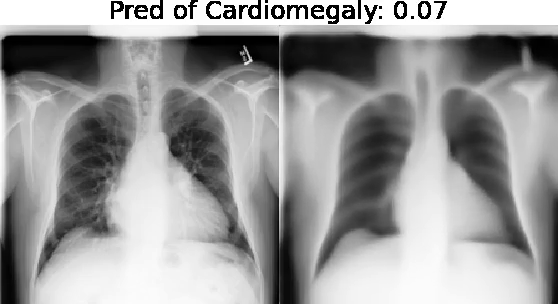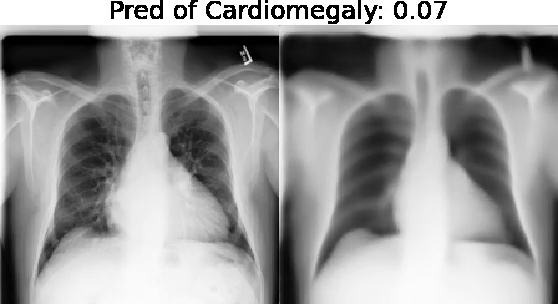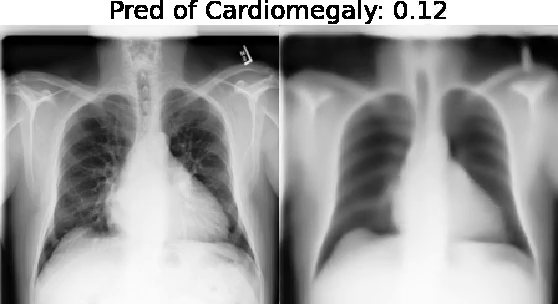
Our approach: Given an arbitrary classifier, we propose a simple autoencoder and gradient update (Latent Shift) that can transform the latent representation of a specific input image to exaggerate or curtail the features used for prediction. We use this method to study chest X-ray classifiers and evaluate their performance. We conduct a reader study with two radiologists assessing 240 chest X-ray predictions to identify which ones are false positives (half are) using traditional attribution maps or our proposed method.
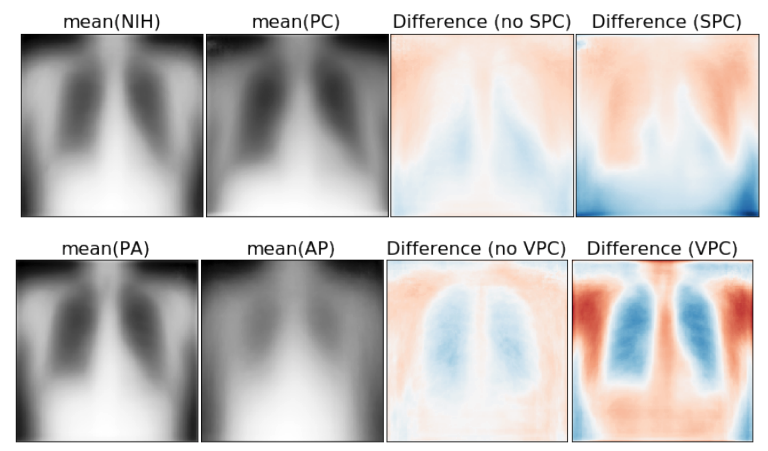
Results: We found low overlap with ground truth pathology masks for models with reasonably high accuracy. However, the results from our reader study indicate that these models are generally looking at the correct features.
We also found that the Latent Shift explanation allows a user to have more confidence in true positive predictions compared to traditional approaches (0.15±0.95 in a 5 point scale with p=0.01) with only a small increase in false positive predictions (0.04±1.06 with p=0.57).